Comparative analysis of polyspecificity of the endogenous tRNA synthetase of different expression host towards photocrosslinking amino acids using an in silico approach |
| |
| Affiliation: | 1. Department of Chemistry, Gottwald Center for the Sciences, University of Richmond, 28 Westhampton Way, Richmond, VA 23173, United States;2. Department of Chemistry, Truman State University, 100 E. Normal Ave, Kirksville, MO 63501, United States;1. Department of Chemistry, Faculty of Science, Kasetsart University, 50 Ngam Wong Wan Rd., Chatuchak, Bangkok 10900, Thailand;2. Department of Social and Applied Science, College of Industrial Technology, King Mongkut’s University of Technology North Bangkok, Bangkok 10800, Thailand;1. School of Science, Xi’an Polytechnic University, Xi’an 710048, PR China;2. School of Mathematics and Statistics, Xidian University, Xi’an 710071, PR China |
| |
| Abstract: | 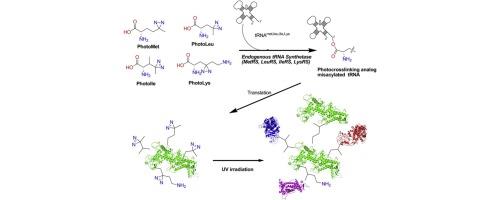
Photo-induced covalent crosslinking has emerged as the powerful strategy for analyzing and characterizing the protein–protein interaction and mapping protein 3D conformations. In the last decades, a number of photocrosslinking amino acids have been reported but only a few have been efficiently utilized for photocrosslinking purposes. Recently, incorporation of diazirine containing photoactivatable analogs such as photo-methionine, photo-leucine, photo-isoleucine and photo-lysine into target proteins were accomplished in live cells (Human A549cells, HEK 293) by depleting corresponding natural amino acid and supplementing these analogs in the medium. Likewise, incorporation of photo-methionine and photo-leucine is also reported in E. coli. Incorporation of these unnatural amino acids were demonstrated only in a limited number species, thereby conventional methods have been utilized for the protein–protein interaction study in other species. With this in mind, we studied in silico analysis of polyspecificity of four endogenous tRNA synthetases (LeuRS, IleRS, MetRS, and LysRS) from six different species such as Escherichia coli, Pseudomonas fluorescens, Corynebacterium glutamicum, Saccharomyces cerevisiae, Aspergillus oryzae and Homo sapiens towards its photocrosslinking amino acids. In addition, here we describe the active site similarity of different protein bio-factories. Based on the active site similarity and similar binding mode, we predicted that the endogenous tRNA synthetases of all the species are reactive to corresponding photoactivatable analogs. This is the first in silico study to demonstrate that the photocrosslinking unnatural amino acids are recognized by the endogenous tRNA synthetases of different protein expression biofactories. |
| |
| Keywords: | Polyspecificity Endogenous tRNA synthetase Molecular docking Substrate specificity Methionine Leucine Isoleucine Lysine Photoanalogs |
| 本文献已被 ScienceDirect 等数据库收录! |
|

